
#VMD SYSTEMS SOFTWARE#
These plugins increases the set of features and tools of VMD making it one of the most used software in computational chemistry, biology, and biochemistry. This communication allows the development of several external plugins that works together with VMD. VMD can communicate with other programs via Tcl/ Tk. In subsequent developments, Jordi Cohen, Gullingsrud, and Stone entirely rewrote the graphical user interfaces, added built-in support for display and processing of volumetric data, and the use of OpenGL Shading Language. In 2001, Justin Gullingsrud, and Paul Grayson, and John Stone added support for haptic feedback devices and further developing the interface between VMD and NAMD for performing interactive molecular dynamics simulations.
#VMD SYSTEMS WINDOWS#
The first version of VMD for the Microsoft Windows platform was released in 1999. In 1998, John Stone became the main VMD developer, porting VMD to many other Unix operating systems and completing the first full-featured OpenGL version. Ulrich in 1995–1996, followed by Sergei Izrailev and J. The earliest versions of VMD were developed for Silicon Graphics workstations and could also run in a cave automatic virtual environment (CAVE) and communicate with a Nanoscale Molecular Dynamics ( NAMD) simulation. The initial version of VMD was written by William Humphrey, Andrew Dalke, Ken Hamer, Jon Leech, and James Phillips. A precursor program, called VRChem, was developed in 1992 by Mike Krogh, William Humphrey, and Rick Kufrin. VMD has been developed under the aegis of principal investigator Klaus Schulten in the Theoretical and Computational Biophysics group at the Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana–Champaign. The VMD axes are shown as a simple example of rendering of non-molecular geometry.
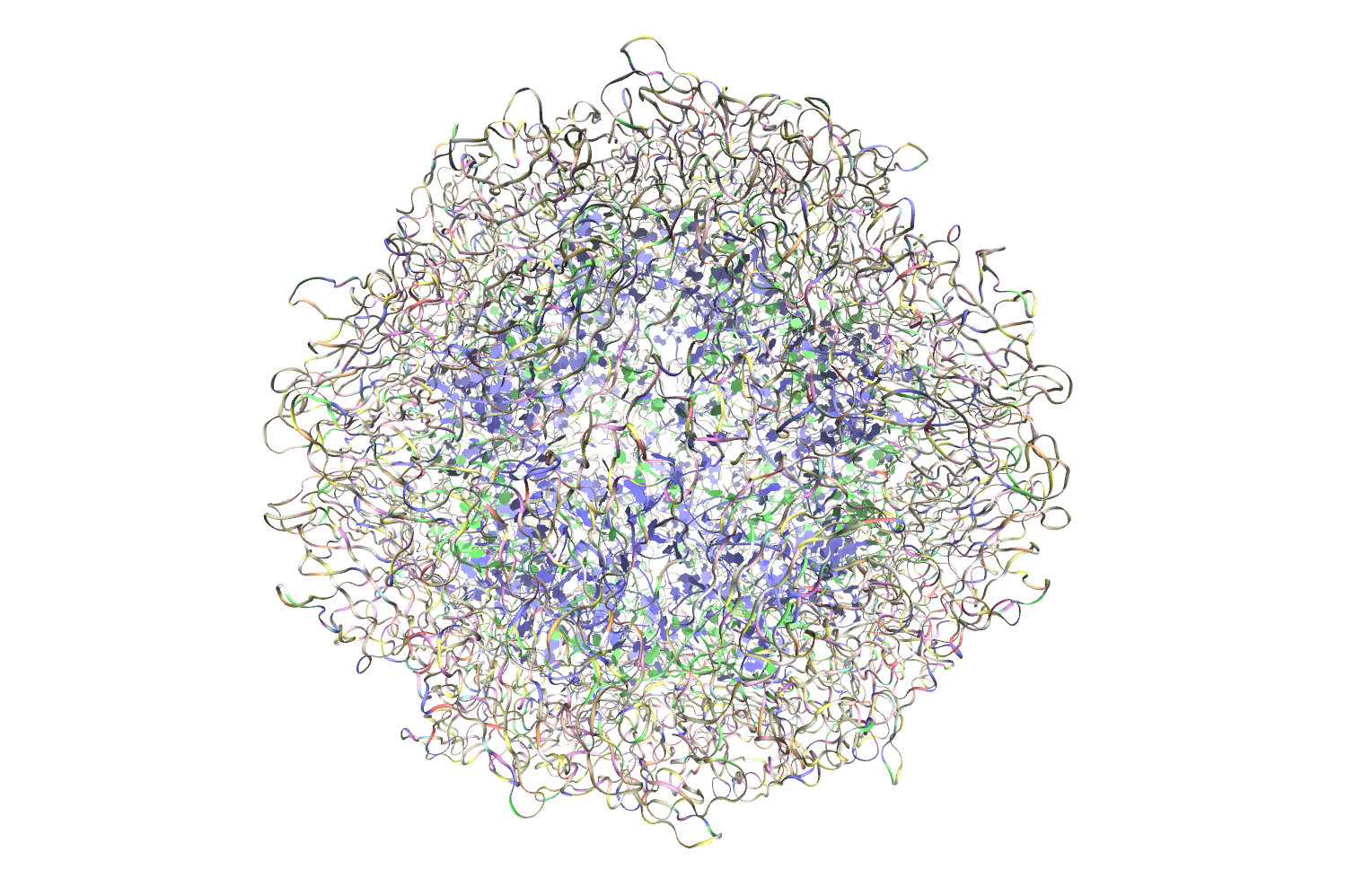
The Tachyon rendering uses both direct lighting and ambient occlusion lighting to improve the visibility of pockets and cavities. The scene is shown with a combination molecular surfaces colored by a radial distance, and nucleic acids shown in ribbon representations. Satellite tobacco mosaic virus molecular graphics produced in VMD and rendered using Tachyon.


 0 kommentar(er)
0 kommentar(er)
